March 2024
(Originally an X thread, dated 9 March 2024)
New thread – new theory
Is this unthinkable?
I think microbiologists have overlooked something huge—something that could have been understood at any time in the last 100 years if the right experiments had been done.
I start with the observation that the error rate when microbial genomic DNA/RNA is replicated must fluctuate due to natural variation.
Moreover, a demand for multiple mutations is created if a microbe is subjected to strong selective pressures.
The quickest way to deliver them may be to start by selecting mutations in the polymerase (those crosses and circles are supposed to be mutations!)
The idea is shown schematically below: consider a hypothetical virus with seven genes – a nucleic acid polymerase, three structural proteins, and three essential non-structural ones. Assume the virus has just spilled over to a new host, and beneficial mutations are required in all three structural proteins to adapt fully. Over time, sequential beneficial mutations may arise, as shown in Panel A, allowing the virus to adapt. If, however, a mutation first arises in the polymerase gene Panel B, a well-adapted strain may arrive much more quickly. However, such a strain would have a high mutation rate, and might be unstable because fatal mutations might arise too rapidly to be removed by “purifying” selection (Panel B).

A proposed lab experiment shown below encapsulates the same idea: under strong selection, microbes may become well-adapted (Series S1) but lose replicative fidelity (shown in Series S2).⬇️
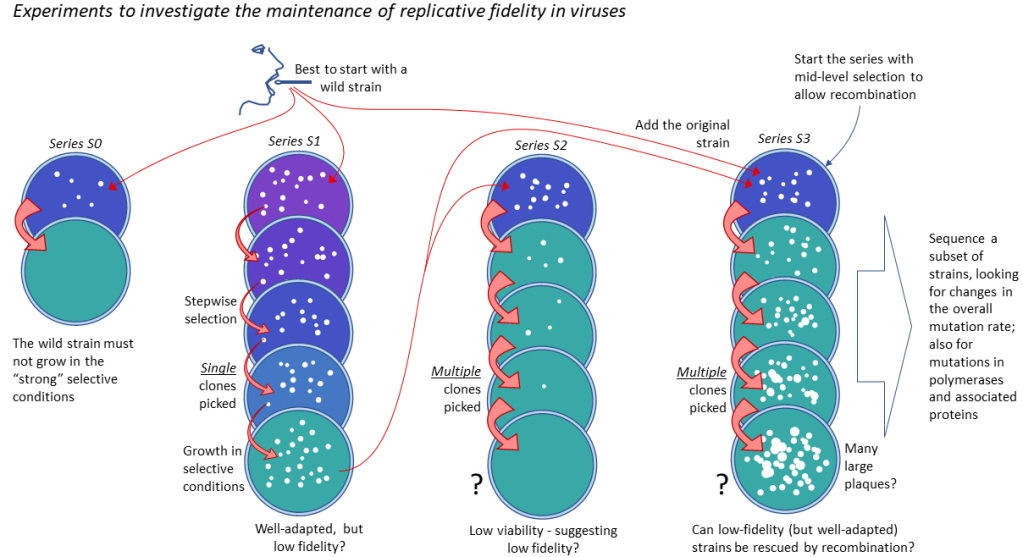
But lineages with low fidelity have limited viability—because mutations will accumulate in essential genes (Series 2).
However . . . cultures containing these well-adapted but low-fidelity strains could, in principle, be “rescued” by recombination with high-fidelity strains – if they are available (see below and Series S3).
I think this happens constantly in nature and in the lab, but microbiologists don’t notice. It’s right in front of them, I suggest, whenever they “stress” their cells, or find one of their engineered constructs doesn’t replicate as strongly as they expected.
[Incidentally, IMO something similar happens in all complex organisms – which is why sexual reproduction is so popular with protists, fungi, plants, and animals – see https://vixra.org/abs/2303.0056 ]
What evidence supports this theory?
Well, first, there’s logic. If you demand a lot of mutations, fidelity is BOUND to fall by natural selection. But since life is still here, fidelity can be reinstated after a fall- which is where recombination, conjugation, etc, must come in.
Second, this proposal can explain why it’s sometimes difficult to propagate and maintain a stable strain of a newly-sampled bacterium or virus in the lab. For example, most archaea have not been isolated, and have been detected only from their gene sequences from environmental samples. Fidelity is often lost (I suggest) early on in the strong selective conditions of a lab, and recombination may not be feasible, making lab propagation difficult or impossible.
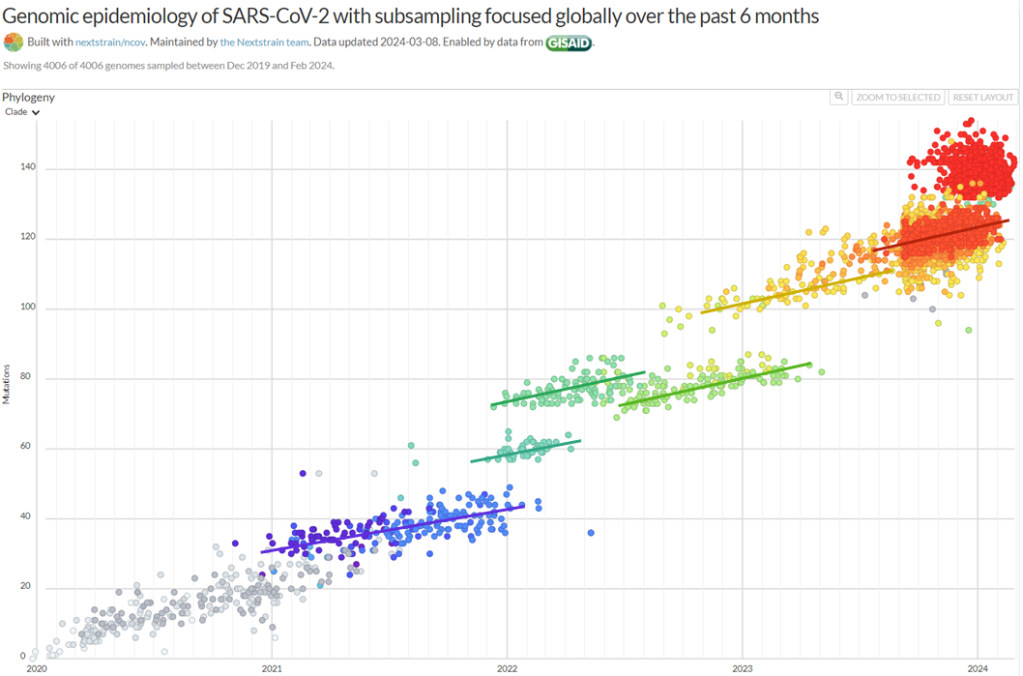
Then, there’s the extraordinary epidemiology of Covid-19. Why did the waves shown below ⬇️ arise so suddenly but then immediately collapse? They lacked the plateaus predicted by theory when immunity builds up slowly. I suggest these strains had slightly lower fidelity, allowing them to evolve reduced immunogenicity, etc., very rapidly but resulting in the subsequent accumulation of mutations in essential genes.

This can also explain how variants such as Delta, Omicron etc evolve. They may be the result of recombination between high- and low-fidelity strains.
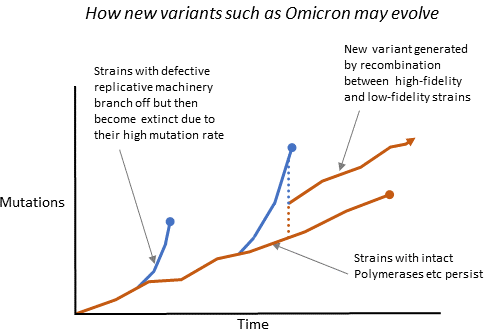
This theory can also explain why some SARS-CoV-2 variants have many mutations in Spike but much fewer in the rest of the genome and why an anomalously low proportion of the mutations in the spike of Omicron (and other variants) were C-to-T nucleotide “transitions”. In high-fidelity strains, most C-to-T transitions come from host modifications of viral RNA, whereas low-fidelity polymerases are expected to generate all nucleotide exchanges randomly.
Finally, we must explain why 100s of thousands of animal viruses can infect and often kill humans, but very rarely cause pandemics. Some, such as Lujo, Omsk, and Rift Valley viruses cause hemorrhagic fevers. Moreover, Marburg, Ebola, and Lassa have all been seen to spread from person to person. Lassa infects about 400,000 people, and kills around 5,000 every year (see Wikipedia entry), but, like the others, has never yet caused a pandemic. In the last 75 years, only seven pandemics have caused more than 10,000 deaths.
This is what I think happens: when an animal virus spills to humans, it rapidly adapts in the first patient and, in the process, develops a high mutation rate. For a pandemic to start, therefore, multiple spillovers need to occur until someone is infected simultaneously by well-adapted and high-fidelity strains, allowing recombination⬇️.
What happened to SARS-1? I guess it developed low fidelity, which recombination couldn’t restore because ongoing animal spillovers were prevented.
Best Origin of CoV-19 theory? SARS-2 was stable and well-adapted when it appeared, so repeated spillovers and recombination probably occurred over years in, e.g., South China. This may have resulted in a high-fidelity human-adapted virus. It seems likely scientists brought the virus to Wuhan, but it could have arrived by another route.
Was it “adjusted” in the lab along the lines suggested in the DEFUSE grant application? Or was a chimera constructed from different human or bat viruses? We don’t know.
So I’m not super-worried about future pandemics – as long as scientists don’t get involved, and as long as we don’t ignore repeated spillovers of animal pathogens in any particular region.
Does anyone know any papers—especially experimental papers —that can shed light on the conservation of replicative fidelity? Let me know.